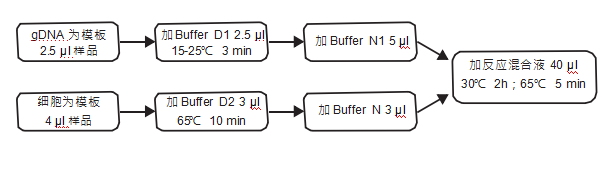
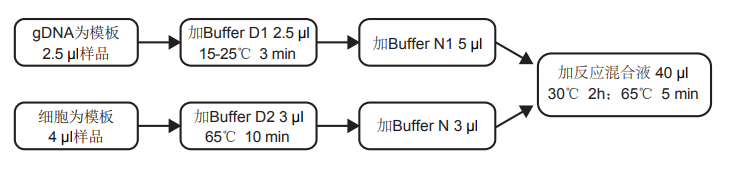
计算溶液所需的质量、体积或浓度。
Single Cell WGA Kit(MDA)
库存信息
库存信息
| 货号 (SKU) | 包装规格 | 是否现货 | 价格 | 数量 |
|---|---|---|---|---|
| S665716-24T |
24T |
期货  |
| |
| S665716-96T |
96T |
期货  |
|
 首页
首页 400-620-6333
400-620-6333